P-type ATPase Motifs
The only thing needed to classify a protein as a P-type ATPase is a 7 amino acid motif next to the aspartate which is phosphorylated during the reaction cycle. The motif can be written as D-K-T-G-T-[LIVM]-[TIS] and it constitutes motif no. PS00154 in the PROSITE database of protein motifs. This motif is present in all currently known P-type ATPases.
The PROSITE motif covers 6 different motifs observed.
DKTGTLT is the most abundant motif and is found in the vast majority of P-type ATPases (currently 356).
DKTGTIT is found in 28 P-type ATPases.
DKTGTVT is found in 3 P-type ATPases.
DKTGTLS is found in 3 P-type ATPases.
DKTGTMT is found in one P-type ATPase.
DKTGTII is found in variants of the human WND / ATP7B Protein, a type IB P-type ATPase.
The above motif allows the classification of proteins as P-type ATPases, it does not allow the classification of P-type ATPases into the different subfamilies. This is a little more difficult, as the P-type ATPases are very divergent. I include below an alignment of the conserved regions from the different types of P-type ATPases. This alignment can also be found in
Axelsen, K.B. & Palmgren, M.G. (1998) J. Mol. Evol. 46: 84-101. It is not always straight forward to determine the type of P-type ATPase, as the motifs are short and not always well defined. Instead it is possible to send protein sequences to me and let me find the type and nearest relatives.
1 25 45 71 95
Segments aaaaaaaaaaaaaaaaaaaaaaaaa bbbbbbbbbbbbbbbbbbbb cccccccccccccccccccccccccc dddddddddddddddddddddddd
All 158/159 ---------------------D--- -------GE----------- -------------------------- ------------P-----------
All 135/159 -------GD------G--iP-D--- ------TGE----------- --------G---------G------- ------------P-----------
All 090/159 -----V-GDIv-v--Gd-iPAD--- vD-S-LTGES-PV-K----- f-G-----G-----V---G--T--G- ----v-i-V--iP--L---v----
IA 3/ 3 ----L---D---V--G--IP-DGE- VDESAITGESAPV--E-G-D TGGT---SD----------G--F-DR ---L-ALLV-LIPTTIG-LLSAIG
IB 23/35 -------GD---V-PG--i--DG-v -D-S--TGE--PV-K--G-- --G--N--G---v--------T---- ------VLvIACPCALGLATP---
IB 28/35 ------------v--G-----DG-v ------TGE--Pv------- --G-----G----------------- ------VLvi-CPC-L----P---
IB 35/35 ---------------------DG-- -------GE----------- -------------------------- ------------P-----------
IIA 17/26 -A-eLVPGDIV---VGD-VPAD-R- vd-S-LTGES--V-K--e-- F-GT----G----vVv-TG--TE-G- F-IAVALAVAAIPEGLPAViT-CL
IIA 21/26 -----VPGD-V----G--vPAD-R- v--S-LTGE---V-K----- f-GT----G----vV--TG--T--G- --i-V--AVA-IPEGLP-ViT--L
IIA 26/26 -------GD-v----G----AD--- -----L-GE---v-K----- f-------G---------------G- -------AV--iPEGLP--iT--L
IIB 8/12 -v-d--VGDI-----GD--PADGv- IDESSLTGESd-v-K--d-- LSGT-V-EGSG-M-VTAVGvNS--G- FII-VTv-VVAVPEGLPLAVT-SL
IIB 10/12 -v----VGDi-----GD--PADG-- iDESS-TGES--v-K----- -SGT-V-eG-G-M-vT-VG--S--G- FI--vTv-VVAVPEGLPLAVT--L
IIB 12/12 -------GDi-----G----AD--- ----S--GE----------- -SG-----G-----v--VG-----G- ----v-v--v-vPE-LPL-v---L
IIC 14/21 -AEieVVGD-VEvKGGDRiPAD-Ri VDNSSLTGESEPQ-RS-efT FFST---EGTA-GiVI-TGD-Tv-GR vIFLIGIIVANVPEGLLATVTVCL
IIC 17/21 -----V-GD-VEvK-GD-iPAD-Ri VDNSSLTGESEPQ-R--e-T FfST---EGT--GiVI--GD-Tv-GR viF-IGIIVANVPEGLLATVTV-L
IIC 21/21 -------G--V----G--iP---R- VD-SSLTGESe-Q------- -------e----G-vi--G-----G- -i-----iVA-VPEG---TvT---
IID 3/ 3 -S---V-GD------GD--PADLRL TDE-LLTGESLPV-KD---- -SSS-V-KGRA-GI---T-----IG- -IYA-----S-IP-SL--VL-ITM
IIIA 21/32 eA--LVPGDIi-i--G-IiPAD-RL iDQSALTGESLPV-K--GD- fSGST-K-GE---VV-ATG--TFFG- ----LV-LI-GIPIA-P-V---TM
IIIA 26/32 -A---VPGDI-----G-iiPAD-R- iDQSA-TGESL-V-K--Gd- fS-S--K-GE---vV-ATG--TF-G- ----L---I-GiPi--P-V---TM
IIIA 32/32 -----V-Gd------G-----D--- iD----TGES---------- ---S----GE----v--T---Tf-G- --------i---P-----V---T-
IIIB 3/ 3 PI--LVPGD---LAAGD--PAD-R- --Q--L-GESLPVEK----- -MGT-V-SG-AQA-V-ATG--TWFG- -LFAL-VAVGLTPEMLPMIV-S-L
IV 7/11 -W--v-VGDiV-i---d-IPAD-iL IET--LDGETNLK-R----- -d--LLRG--L-NT--v-GiVv-TG- ---fvILf---vPISL-V--E-iK
IV 9/11 ----v-VGD-v-------iPAD-iL i-T--LDGET--K------- ----------L-------G-Vv--G- -----I-f---vPISL-v--e--K
IV 11/11 -------GD-v---------AD--- i-T--LDGET--K------- --------------------V---G- -----------vP--L-v-----K
V 5/ 7 ---ELVPGDI---------PCD-iL V-E-MLTGESVPi-K----- --------------ViRTGF-T-KG- I---LDIIT--vPP-LP----v-i
V 6/ 7 ------P-DI---------PCD--- V-E--LTGESVP--K----- --------------V--TGF-T--G- ------I-T--vPP-LP-------
V 7/ 7 --------D------------D--- --E----GE-----K----- --------------V--T-f----G- ------I-----PP--P-------
96 131 188
Segments eeeeeeeeeeeeeeeeeeeeeeeeeeeeeeeeeeee fffffffffffffffffffffffffffffffffffffffffffffffffffffffff
All 158/159 ---------------------DKTGT---------- ----------------------------------------GD---------------
All 135/159 ----------E----------DKTGTLT-------- ---------------D---------i------G------TGD----A-------G--
All 090/159 ---V----AvE-LG----iCSDKTGTLT-N---V-- ---------G-----DPPR------i------GI-V-MiTGD---TA-AIA---Gi-
IA 3/ 3 NV-ATSGRAVEA-GD---L-LDKTGTITLGNR-A-- ---------GV--LKDI-K-GI-E-F---R-M---TVM-TGDN--TA--IA-EAGVD
IB 23/35 GiLiK----LE-------v-FDKTGTLT-G---V-- ------------Av-D---------i--L---G-----LTGDN---A-AiA---GI-
IB 28/35 GiL-K----LE-------v--DKTGTLT-G---V-- ---------------D---------i--L---G------TGD----A--iA---G--
IB 35/35 ---------------------DKTGT---------- ----------------------------------------GD---------------
IIA 17/26 NAiVR-LPSVETLGC--VICSDKTGTLTTN-M-V-- --E--L---G-vG--DPPR-EV--AI--C--AGIRV-MITGD---TA-AI---IG--
IIA 21/26 -A--R-LP-VETLG---VICSDKTGTLT-N-M-V-- -----L---G--G--DPPR--V---i--C--AGI-v--ITGD---TA-AI----G--
IIA 26/26 ------L--VE-LG----iC-DKTGTLT---M---- ---------G-----D--R--------------i-----TGD---TA--i-------
IIB 8/12 NNLVRHL-ACETMG-AT-ICSDKTGTLT-N-MTVV- -----LT-I--VGI-DPVRPEVP-A---C--AGITVRMVTGDNI-TARAIA--CGIL
IIB 10/12 -NLVR-L--CETMG-AT-ICSDKTGTLT-N-M-VV- ------------GI-DP-R--V--A---C--AGI-VRMVTGDN--TA-AIA--C-I-
IIB 12/12 --LVR----CETM------C-DKTGTLT-N-M---- ------------GI-D--R--V------C--AG--VRMV-GDN--TA--IA--C-I-
IIC 14/21 NCLVKNLEAVETLGSTS-ICSDKTGTLTQNRMTVAH FP---LCFVGL-SMIDPPRA-VPDAV-KCRSAGIKVIMVTGDHPITAKAIAK-VGII
IIC 17/21 NCLVKNLEAVETLGSTS-ICSDKTGTLTQNRMTVAH FP---L-F-GL-SMIDPPR--VPDAV-KCRSAGIKVIMVTGDHPITAKAIA--VGII
IIC 21/21 ------L--vETLGS---I-SDKTGTLTQNRMTV-H F------F--L-S---PPR--V--AV--C---GI-ViMVTGDHPITA-AIA--V-II
IID 3/ 3 -V-VR-L--LEALG-V-DICSDKTGT-TQG-M--R- --E--L-F--L-GIYDPPR-E--GAV---H-AGI-VHMLTGD---TAKAIA-EVGI-
IIIA 21/32 -AI-----AIEE-AG-dvLCSDKTGTLTLNKLSv-- ---G-W---G--P--DPPRHD-AeTI--A--LGv-VKMiTGD---I-KET-R-LGMG
IIIA 26/32 -AI-----AIE--AG-dvLCSDKTGTLT-NKL---- -----W---G--P--DPPR-D---TI--A--LG--VKM-TGD---I-KET-R-LG-G
IIIA 32/32 --I------iE-------LC-DKTGTLT-N------ -----W---------DPPR-D----i------G--vKM-TGD---I-------L---
IIIB 3/ 3 KVIVK-L-AIQNFGAMD-LCTDKTGTLTQD-I-LE- -DE--L--EG---FLDPPKE----A--AL---G--VK-LTGD---V-A--C-EVG-D
IV 7/11 ----R---I-EELGQ-EYIFSDKTGTLT-N-M-FKK -IE--L-LLG-T-IED-LQDGV-d-IE-L--AGIKiW-LTGDK-ETAINIG-S--L-
IV 9/11 ----------eeLG---Yi-SDKTGTLT-N-M-F-K --E--L-L-G-T-iED-LQ--V------L--AGI--W-LTGDK-ETA--I--S--L-
IV 11/11 -----------eLG-------DKTGTLT-N-M---- --E----L---T-iED-LQ--V---------A-I--W-LTGD--ETA--i-------
V 5/ 7 -IfC--P--i---G-I---CFDKTGTLTED-L---G --E--L-FLGF-i--N-LK--T---I--L-------iMiTGDN-LT---V--E-GIi
V 6/ 7 -I-C--P------G-I---CFDKTGTLT-d-L---- --E--L---G--i-----K------I-----------M-TGDN-LT---V------i
V 7/ 7 -i----P------G-i----FDKTGTLT-------- --E------G--i-----K---------------------GDN--T---v-------
234 265
Segments gggggggggggggggggggggggggggggggggggggggggggggg hhhhhhhhhhhhhhhhhhhhhhhhhhhhhhh
All 158/159 ------P--K-----------------GDG-ND------------- -------------------------------
All 135/159 ------P--K---v-------------GDG-ND-P-L--A-iG--- ---------d---------------------
All 090/159 vFA---P--K--iV--LQ-----VAMTGDGVNDAPALK-ADiGIA- GTDvA--AAD-vL-d-----Iv-Av---R-i
IA 3/ 3 F-AEATPE-K---I---Q--G-LVAMTGDGTNDAPALAQA-V--AM GTQAA-EA-NMVDLDS-PTKLI--V-IGKQ-
IB 23/35 v-A---P-dK---i--L---G--VAMVGDGINDAPALA-A--GiA- GTDvA---ADi-L----L--v-----L---T
IB 28/35 ------P--K---i---------V-MVGDGiNDAP-LA-A--G-A- G-DvA---AD--L----L-------------
IB 35/35 ------P--K-----------------GDG-ND------------- -------------------------------
IIA 17/26 -F-R--P-HK--IV--L----ei-AMTGDGVNDAPALK-AdIGIAM GT-VAK-ASdMVL-DDNF-TIV-AV-EGR-I
IIA 21/26 -F-R--P-HK---V--L-------AMTGDGVNDAPALK-AdIGiAM GT-VAK-A-dMvL-DDNF-TIV-Av-EGR-I
IIA 26/26 ---R-----K---v----------AMTGDG-N---AL--------- ---V--------L-DD-F--i--A---G--i
IIB 8/12 VLARSSP-DK-TLV----d---VVAVTGDGTND-PALK-ADVGFAM GTdVAKEASDIIL-DDNF-SIV-AV-WGRNV
IIB 10/12 V-ARS-P-DK--LV--------VVAVTGDGTND-PALK-ADVG-AM GTdVAKeASDiI--DDNF--IV--v-WGR-V
IIB 12/12 V-AR--P-DK------------vVAVTGDG-ND-PAL-----G--M GT-VA-----ii---D-F--IV------R-v
IIC 14/21 VFARTSPQQKLIIVEGCQR-GAIVAVTGDGVNDSPALKKADIGVAM GSDVSKQAADMILLDDNFASIVTGVEEGRLI
IIC 17/21 VFARTSPQQKLIIVEGCQR---iVAVTGDGVNDSPALKKADIGvAM GSD--K-AADMILLDDNFASIVTGVEEGRLI
IIC 21/21 VFARTSP-QK--IVE--Q-----V-VTGDG-ND-PAL-KADIGvAM G-D--K--AD-iLL-DNFAS-V-GvE-GR-I
IID 3/ 3 VI-RC-PQTKV-MIEALHRR--F--MTGDGVNDSPSLK-ANVGI-M GSDV-K-ASDIVL-DDNF-SI-NA-EEGRRM
IIIA 21/32 GFA-VFPEHKY-IV--LQ-R---V-MTGDGVNDAPALKKAD-GIAV ATDAAR-A-DIVLT-PGLS-II-A--TSR-I
IIIA 26/32 GFA-VFP-HKY-iV--LQ-------MTGDGVNDAP-LKKAD-GIAV ATDAAR-A-DIV---PGLS-II-A---SR-I
IIIA 32/32 GF----P--Ky--------------MTGDGVND-P-LK-A--GiA- --DAAR---D-V----G---ii-A----R-i
IIIB 3/ 3 -FARLTP--K-RI---L---GH-VGF-GDGINDAPALR-AD-GISV A-DIA-E--DIILLEK-LMVLEEGVI-GR-T
IV 7/11 iCCR-SP-QKA-vV--v-------LAIGDG-NDV-MIQ-A-VGvGI -EG-A----DY-I-QF--L--LLLVHGR--Y
IV 9/11 -CCR--P-QKA-vV--v--------AIGDG-NDV-MIQ-A-vGvGI -EG-A----Dy-I-QF-----L-LVHG---Y
IV 11/11 ---R--P--K----------------I--G-ND--MI--A--GvGI -EG-A----D-----F-----L---HG---Y
V 5/ 7 VyARM-P-QK--Li--L------V-MCGDGANDC-ALK-A-vGISL ---eASiAAPFTS----i--V--VI-EGR--
V 6/ 7 VyAR--P--K------L--------MCGDG-ND--AL--A--GI-L ---e-S----FTS-------v--v---GR--
V 7/ 7 vyAR--P--K------L----------GDG-ND--A---A--Gi-- ---e------f-S-------v--v---GR--
The fingerpritns here are based on the P-type ATPases included in the phylogenetic analysis performed in
Axelsen, K.B. & Palmgren, M.G. (1998) J. Mol. Evol. 46: 84-101. Repetitive lowercase letters on top indicate name of conserved segment. Amino acids identical in all members of given type of P-type ATPases (Table 1) are in bold. The consensus amino acids shown in italic lowercase letters are positions where the sequences have one of two conserved amino acids I or V; D or E; F or Y. Numbers at top indicate the position of amino acids when conserved segments are arranged in a linear core sequence. Consensus sequences at top indicating amino acids present in 158, 135, and 90 sequences, respectively, are shown.
If the alignment is not formatted perfectly by your browser try to click here.
Most of the type IB P-type ATPases contain an additional motif, the HMA domain believed to consitute metal binding sites in these proteins. More information about this domain is found here.
The type IIC P-type ATPases harbours an additional subunit necessary for the function of this subfamily. These 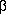 -subunits are recognized by another PROSITE pattern specific for this group of proteins. More information about the
-subunits are recognized by another PROSITE pattern specific for this group of proteins. More information about the 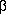 -subunits is found here.
-subunits is found here.
Go to Front page
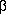 -subunits are recognized by another PROSITE pattern specific for this group of proteins. More information about the
-subunits are recognized by another PROSITE pattern specific for this group of proteins. More information about the 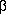 -subunits is found here.
-subunits is found here.